BRAIN
Probing Fine-scale Connections in the Brain
Esther Landhuis
Nature | October 19, 2020
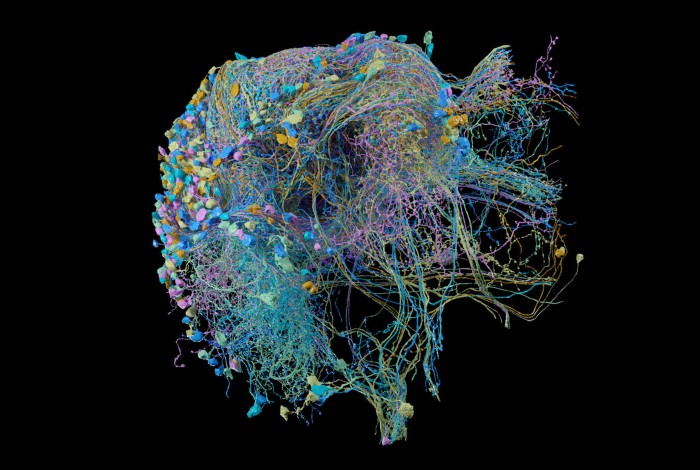
Tracing connections, such as those in this section of the fruit-fly brain, could uncover links between neural architecture, biology and disease. Credit: FlyEM at HHMI/Google Research
There are 70 million neurons in the mouse brain, and Moritz Helmstaedter wants to map them all. He was a medical student at Heidelberg University in Germany when psychiatrists there suggested that some aspects of the human psyche lack a biological explanation. “I was totally appalled,” recalls Helmstaedter, who is now a director at the Max Planck Institute for Brain Research in Frankfurt, Germany.
Although the brain remains a mystery, Helmstaedter was convinced that what goes on there “must be a mechanistic phenomenon in the end, as complex as it may be”. He has dedicated the past two decades to working those mechanisms out — and he and other neuroscientists are finally starting to scratch the surface, one cubic micrometre at a time.
Starting in the 1970s, it took more than a decade to unravel the neural circuitry of the one-millimetre worm, Caenorhabditis elegans. Probing the relationship between genes and behaviour, biologist Sydney Brenner and his colleagues at the MRC Laboratory of Molecular Biology in Cambridge, UK, laboriously traced the fine branches and synaptic connections of each nerve cell, colour-coding them by hand on thousands of electron-micrograph prints. That wiring map — the first and only complete set of synaptic connections in an animal’s nervous system — was stored on a room-sized computer and published1 as the first full animal ‘connectome’ in a 340-page opus in 1986.
Caenorhabditis elegans has fewer than 400 neurons; human brains have 86 billion. So for now, scientists are eyeing an intermediate milestone: mapping the fine-scale neural circuitry of the mouse2.
Even with about 1,000-fold fewer cells, the mouse brain poses a formidable challenge, says Jeff Lichtman, a neuroscientist at Harvard University in Cambridge, Massachusetts, who is one of the leaders of a global consortium that aims to reconstruct the neural wiring of a mouse brain over the next decade. “We’re dealing with a data set that will be on the scale of an exabyte.” An exabyte is one billion gigabytes; the entire human genome can be represented in about 1.5 gigabytes. In terms of data size, mapping the mouse brain connectome will be “enormous compared to anything that’s been done as a single project”, he says. “Connectomes are just magnificently complicated.”
Yet the technology to make such an undertaking possible is nearly there. With advances in microscopy and artificial intelligence (AI), and crowdsourced help from human gamers, researchers are beginning to map neural networks and their connections at ever-higher resolution and scale. Over the past several years, small bits of brain, including pieces of the mammalian retina and cerebral cortex, have come into focus. And in September, researchers working on Drosophila fruit flies reported3 the largest reconstruction so far: 25,000 neurons in the hemibrain, a cube of tissue measuring 250 micrometres on a side and representing 40% of the fly’s brain.
These are not mere exercises in big biology. As connectomics pushes the technological and computational limits, researchers hope to tap these data sets to learn how experiences are stored in the brain, with potential insights into autism, schizophrenia and other ‘connectopathies’.
Early developments
After the C. elegans neural-wiring diagram launched connectomics in 1986, the field went silent, says Helmstaedter. It was an issue of technology: researchers had no way, beyond what Brenner’s team had done, to probe neural circuits at connectome scales.
As a doctoral student in the early 2000s, Helmstaedter stuck electrodes into nerve cells to figure out which ones were electrically connected, an approach that might allow the simultaneous recording of four or five neurons. Yet networks have hundreds or thousands of nerve cells and millions of connections. “To really map the circuits, we needed something else,” he says.
That came in 2004. Winfried Denk, then at the Max Planck Institute for Medical Research in Heidelberg, and his colleagues installed a precision-cutting tool called a microtome in the vacuum chamber of an electron microscope (EM), making it possible to automate nanoscale imaging. It revitalized the field4.
Called serial block-face scanning electron microscopy (SBEM), Denk’s method involves loading a block of tissue into the machine, which then automatically images the exposed face, scrapes off the top layer of tissue and repeats, for days or weeks at a time. In 2013, Denk’s team, led by Helmstaedter, a former postdoc in the lab, used SBEM to map a complete set of synaptic connections for 950 neurons in the mouse retina5. This was a significant undertaking: the cost, including equipment, salaries and some €300,000 (US$350,000) in fees paid to undergraduate students to trace circuits throughout the EM data sets, totalled around €2 million. And it revealed new cell subtypes. But beyond that, the work provided a comprehensive map for researchers to identify interaction partners for cells of interest, Helmstaedter says — “like using a street map for navigation versus trial and error”.
Worm maths
The students who traced those neural circuits did so using computers. That shift began in the early 2000s, when researchers began adopting a computational approach to mapping the connectome. This wasn’t machine learning; humans still did the work. But rather than tracing neurons on paper with coloured pencils as Brenner’s team had, they mouse-clicked through stacks of digitized images.
Biologist Scott Emmons at the Albert Einstein College of Medicine in New York City and his team, for instance, digitized Brenner’s original images and used a computational approach to map the circuits that regulate mating behaviours in the tail of a male C. elegans. (The 1986 effort focused on the other C. elegans sex, the hermaphrodite.)
Connectome of the adult Caenorhabditis elegans brain.Credit: D. Witvliet et al./bioRxiv
Emmons then hired a statistician to apply graph theory — a branch of mathematics used to analyse networks such as bus routes or maps of the spread of disease — to calculate the strength of synaptic connections and work out which parts of the connectome network drive specific behaviours. Although mathematicians had studied artificial neural networks for decades, the C. elegans work, reported6 in 2012, was considered the first quantitative analysis of a natural neural network, Emmons says.
Extending those efforts, in 2019 Emmons and his colleagues published7 quantitative connectomes of the complete nervous systems of both C. elegans sexes, comprising 687 neurons. One goal was to determine how connectivity differed between the male and the hermaphrodite. But distinguishing sex-based differences from natural biological variability among individuals proved a tough nut to crack, delaying the paper for a full year, and requiring still more mathematics. In the end, the team concluded that up to 30% of neural connections might reflect true differences based on sex, Emmons says, as opposed to disparities arising from rearing conditions or methodological errors.
Unexpected complexity
Tracing neural circuitry through the brain of a tiny worm requires thousands of 30–50-nanometre sections. “Even if you’re a god, you cannot cut 5,000 sections without messing anything up,” says Mei Zhen, a neuroscientist at the University of Toronto in Canada.
Now try doing that eight times. In an analysis described in a bioRxiv preprint in May8, researchers led by Zhen, Lichtman and Harvard physicist Aravinthan Samuel reconstructed connectomes across eight developmental stages, to learn how the C. elegans wiring diagram changes as the worm matures from early larva to adult. They sectioned most of their samples with an automated tape-collecting ultramicrotome, developed in Lichtman’s lab, that collects serial sections and places them in sequence on a reel for subsequent imaging by a scanning electron microscope9. It “turns a worm into a roll of tape”, Zhen says.
Using it, the team detected enormous differences in the worms’ wiring. Even among genetically identical animals, Zhen says, about 43% of connections were not the same.
At the MRC Laboratory of Molecular Biology, systems neuroscientist Marta Zlatic studies another wormlike creature: Drosophila larvae. Measuring 4–8 millimetres in length, the larvae are larger than C. elegans but have 10–20-fold fewer brain neurons than adult flies. “So we can reconstruct circuits a lot quicker, and we can do so in many individuals,” says Zlatic, who combines connectomics with other techniques to correlate neural structure with function.
In a study10 published in March, Zlatic’s team analysed 102 neuron pairs involved in odour avoidance. They found that even in these tiny insects, far-flung cellular networks regulate learning. “When an animal learns a new thing about an odour, how quickly and how well it learns that will be influenced by everything else it has ever learnt about that odour,” she says.
Machine learning
Scaling up such analyses to animals such as adult Drosophila or mice, which are capable of higher-order learning, is much more challenging. But there, too, progress is being made.
In one 2019 study11, Helmstaedter and his colleagues used SBEM to image and reconstruct a speck of mouse brain in a region that processes sensory input. Measuring 500,000 cubic micrometres, the volume contained some 2.7 metres of neuronal wiring and 400,000 synaptic connections. It held just 89 neurons, but was 300 times larger than previous reconstructions from the mammalian cerebral cortex.
Reconstructing these circuits with then-standard techniques — moving from slice to slice, manually tracing each nerve cell — would have taken hundreds of thousands of hours, Helmstaedter estimates. So, his team combined automated image-processing algorithms with machine-learning AI approaches, and focused human effort on marking neuron branches while letting computers handle the volumetric reconstruction. This cut the workload to 20,000 hours — still the equivalent of 10 people working full-time for a year. Further AI improvements sped up the process still more, by training computers to evaluate the machine-assembled reconstructions and requesting human help only when needed.
Meanwhile, researchers at the Janelia Research Campus of the Howard Hughes Medical Institute (HHMI) in Ashburn, Virginia, set their sights on Drosophila. A fruit fly’s brain has many fewer neurons than a mouse brain, yet its wires are thinner and more densely packed, says Shan Xu, an applied physicist at Janelia. Tracing these connections to completion would require 8-nm resolution — 3–6 times what was possible with scanning EM when Xu joined the HHMI in 2009. Given the technology available then, he says, the imaging alone would have taken a decade.
And that wasn’t even the biggest challenge. To track each neuronal process at this resolution, the images had to be perfect; a 100-nm gap could render the data set useless. “You’d want to operate a machine for a decade with no error,” Xu says.
Xu spent several years painstakingly investigating failure modes and conditions that triggered them. “Before it fails, what are the telltale signs? What are the indicators I can capture and immediately stop the operation? And then how can I restart from there smoothly?” he asked. Instead of perfection, he opted for intelligence, building a system that could shut itself off when a computer glitch, vacuum-pump failure or other malfunction looms. That shift in mindset “completely changed the picture”, says Xu. It helped his team refine another block-face technology, FIB-SEM, that cuts using a focused beam of ions rather than a microtome and produces higher-resolution pictures with few defects12.
Xu and his colleagues ran two of these FIB-SEM systems in parallel for about two years. Then, after a computer aligned the stacks of EM images, a team of 50 HHMI employees laboured full-time for a year to proofread them. Poring through thousands of images per day, proofreaders quickly learnt to spot errors — for example, improperly joined segments or orphaned pieces that needed to be connected. “You develop your eye for the type of neuron you’re looking at,” says Erika Neace, an analytics developer and veteran proofreader at Janelia. In all, the team traced 25,000 neurons and their 20 million connections in the hemibrain3.
Separately, another team at Janelia imaged the fly brain using a transmission electron microscope (TEM), an older technology that passes electrons through thin specimens rather than scanning a surface as FIB-SEM does. Fitting the microscope with an array of cameras and robotic components to process larger specimens at higher speeds, they imaged an entire adult Drosophila brain13. They then validated the resulting data set — all 106 terabytes of it — by meticulously tracing circuitry in the mushroom body, a brain structure important for learning and memory.
Today, researchers are crowdsourcing the annotation process, creating online games as test beds for fine-scale connectomics using AI and human volunteers. In August, researchers led by neuroscientists Sebastian Seung and Mala Murthy at Princeton University in New Jersey released a game-like platform called FlyWire, which presents AI-identified pieces of neurons from Janelia’s fly-brain data set for players to assemble14. Building on Eyewire — an earlier game developed by the Seung lab using a smaller set of mouse-retina images — FlyWire supplements Google’s Neuroglancer visualization platform with tools that let multiple users view and edit neurons in the same data set at the same time.
Unlike Xu’s hemibrain reconstruction, which was processed by an internal proofreading team, FlyWire invites contributions from everyone. And those contributions improve the system over time, says Seung. When a user edits a neuron by clicking a button to split or merge pieces, that action sends a message that trains the machine to detect errant reconstructions and, over time, to correct them. “This is a really profound, philosophically interesting moment,” says Lichtman. “Machines are learning to be smarter by studying the wiring of machines that are fundamentally smarter — biological machines.”
Seung is now collaborating with researchers at Baylor College of Medicine in Houston, Texas, and the Allen Institute for Brain Science in Seattle, Washington, to build a new online community, Pyr (as in, ‘Pyr into the brain’), to map an even larger piece of the mouse brain. In October, the Allen Institute team, led by Clay Reid and Nuno da Costa, described the method used to collect those data. Using six 1980s-era TEMs retrofitted with large-field-of-view cameras and a custom reel-to-reel sample-handling system, the team imaged one cubic millimetre of mouse visual cortex in 6 months, yielding 2 petabytes of data15. Now the team is looking to scale their operations to the whole-brain level, says Wenjing Yin, an Allen Institute scientist who helped to develop the imaging pipeline. “There are a lot of questions and problems we need to solve,” she says.
With some 70 million mouse neurons still uncharted, the connectome community will have its work cut out. But Helmstaedter is optimistic. “It’s an extremely exciting time for charting new territory,” he says, “with all the surprises this always entails.”
Nature 586, 631-633 (2020)
References
- 1.
White, J. G., Southgate, E., Thomson, J. N. & Brenner, S. Phil. Trans. R. Soc. Lond. B 314, 1–340 (1986).
- 2.
Abbott, L. F. et al. Cell 182, 1372–1376 (2020).
- 3.
Scheffer, L. K. et al. eLife 9, e57443 (2020).
- 4.
Denk, W. & Horstmann, H. PLoS Biol. 2, e329 (2004).
- 5.
Helmstaedter, M. et al. Nature 500, 168–174 (2013).
- 6.
Jarrell, T. A. et al. Science 337, 437–444 (2012).
- 7.
Cook, S. J. et al. Nature 571, 63–71 (2019).
- 8.
Witvliet, D. et al. Preprint at bioRxiv https://doi.org/10.1101/2020.04.30.066209 (2020).
- 9.
Baena, V., Schalek, R. L., Lichtman, J. W. & Terasaki, M. Methods Cell. Biol. 152, 41–67 (2019).
- 10.
Eschbach, C. et al. Nature Neurosci. 23, 544–555 (2020).
- 11.
Motta, A. et al. Science 366, eaay3134 (2019).
- 12.
Xu, C. S. et al. eLife 6, e25916 (2017).
- 13.
Zheng, Z. et al. Cell 174, 730–743 (2018).
- 14.
Dorkenwald, S. et al. Preprint at bioRxiv https://doi.org/10.1101/2020.08.30.274225 (2020).
- 15.
Yin W. et al. Nature Commun. 11, 4949 (2020).
